SMDB
SMDB database had been deposited in https://github.com/taylor19891213/sulfur-metabolism-gene-database on January 8th, 2020. The SMDB website (http://smdb.gxu.edu.cn/) had been online since June 22th, 2020.
This section is divided into:
1.Update details
2.Data source and inclusion criteria
3.Database schema
4.Database Structure and Creation workflow
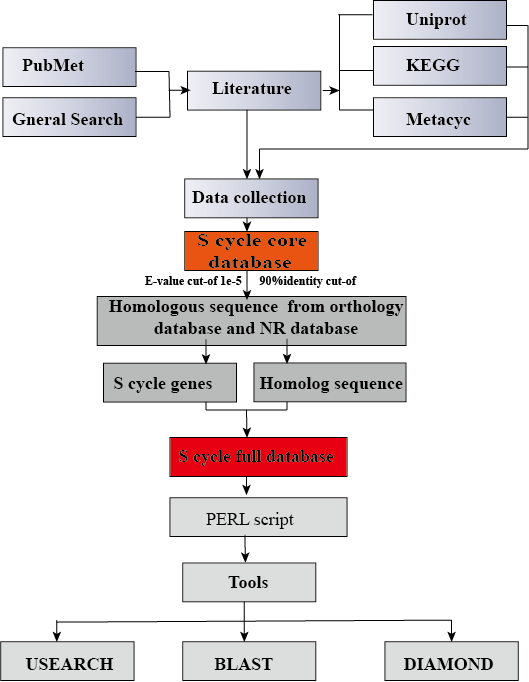
The sulfur cycle is one of the most important biogeochemical cycles in earth ecosystems. Characterizing sulfur metabolism at a gene- and species-level resolution has become an important approach in understanding sulfur metabolism processes in the current metagenomic era. However, a professional and efficient gene integration database of sulfur metabolism has not been established yet. we created a manually managed database that gathers most of the publicly available sulfur cycle gene families to address the limitations of available public databases in sulfur cycle analysis. A total of 175 gene (sub) families covering 11 sulfur metabolism pathways, including assimilatory sulfate reduction, thiosulfate disproportionation, sulfide oxidation, dissimilatory sulfate reduction, sulfite oxidation, sulfur oxidation, sulfur reduction, tetrathionate oxidation, tetrathionate reduction, thiosulfate oxidation, and organic degradation/synthesis, were recruited in SMDB. SMDB obtained 115,321 and 395,737 representative sequences at 95% and 100% identity cutoffs, respectively. The 95% identity cutoff was chosen for its approximately corresponding to the species definition in genomic era. Representative sequences were then selected to construct the full SMDB. So there are two databases namely SMDB.100.fa and SMDB.95.fa, respectively.
1 Update details
This version (version 1) of SMDB database was last updated on May 9, 2019. The database is updated occasionally through formalized literature searches.2 Data source
An extensive review of the scientific literatures related to sulfur cycle is the foundation of SMDB. The SMDB construction utilized the UniProt database (http://www.uniprot.org/) for retrieving core database sequences of sulfur cycle gene families by using keywords. All data have been manually scrutinized before included into SMDB.
Other databases were used for retrieving nontarget homologous sequences. The other databases used included COG (ftp://ftp.ncbi.nih.gov/pub/COG/COG2014/data), eggNOG(http://eggnogdb.embl.de/download/eggnog_4.5/), KEGG (http://www.genome.jp/kegg/), and M5nr (ftp://ftp.metagenomics.anl.gov/data/M5nr/current/M5nr.gz). We also added the homologous sequence of RefSeq non-redundant proteins (NR) database(ftp://ftp.ncbi.nlm.nih.gov/blast/db/. These databases were selected, because they are widely used in metagenome research. SMDB included sulfur cycle gene families and their nontarget homologous gene families. A multistage filtering was performed to collect homologous sequences. Initially, a fixed e-value cutoff of â 0.00001 followed by a fixed coverage cutoff of 90% was used for all sequence matches. Redundancy was removed by clustering all homologous protein sequences with a 100% identity cut-off using USEARCH (version 5.2.32).
3 Database schema
SMDB databases contains two files. One of a file contains the gene sequence, and the other is a file that matches the gene sequence to the its name. (The database are stored in MySQL server 5.7.24 relational databases and operated in CentOS Linux systems. The web-interface was created using HTML, PHP and Cascading style sheets (CSS). PHP programming language was used for retrieving data from MySQL databases and present result output to the web-interface.)
4 Database Structure and Creation workflow